News
(February 2024) Congratulations to Federica for successfully defending her PhD!
Huge congratulations to Federica Capraro for passing her PhD viva with flying colours! Both examiners were incredibly impressed by Federica and her thesis, a magnum opus on LARP6! Of course we didn't expect anything otherwise from our star PhD student ! Well done Dr Capraro! A big milestone in an amazing scientific career to come! We can't wait to see all the great scientific breakthroughs that you are going to make in the future!
A big thank you to our fantastic viva examiners, Professors Anne Willis (University of Cambridge) and Andres Ramos (University College London), for coming all the way to BCI and conducting the viva! It was a great pleasure to have you here and Federica really enjoyed the experience!
Last but not least, massive thanks to Emilie, Zeinab, and other members of Mardakheh and Conte labs for organising a memorable post-viva celebration party for Fede!
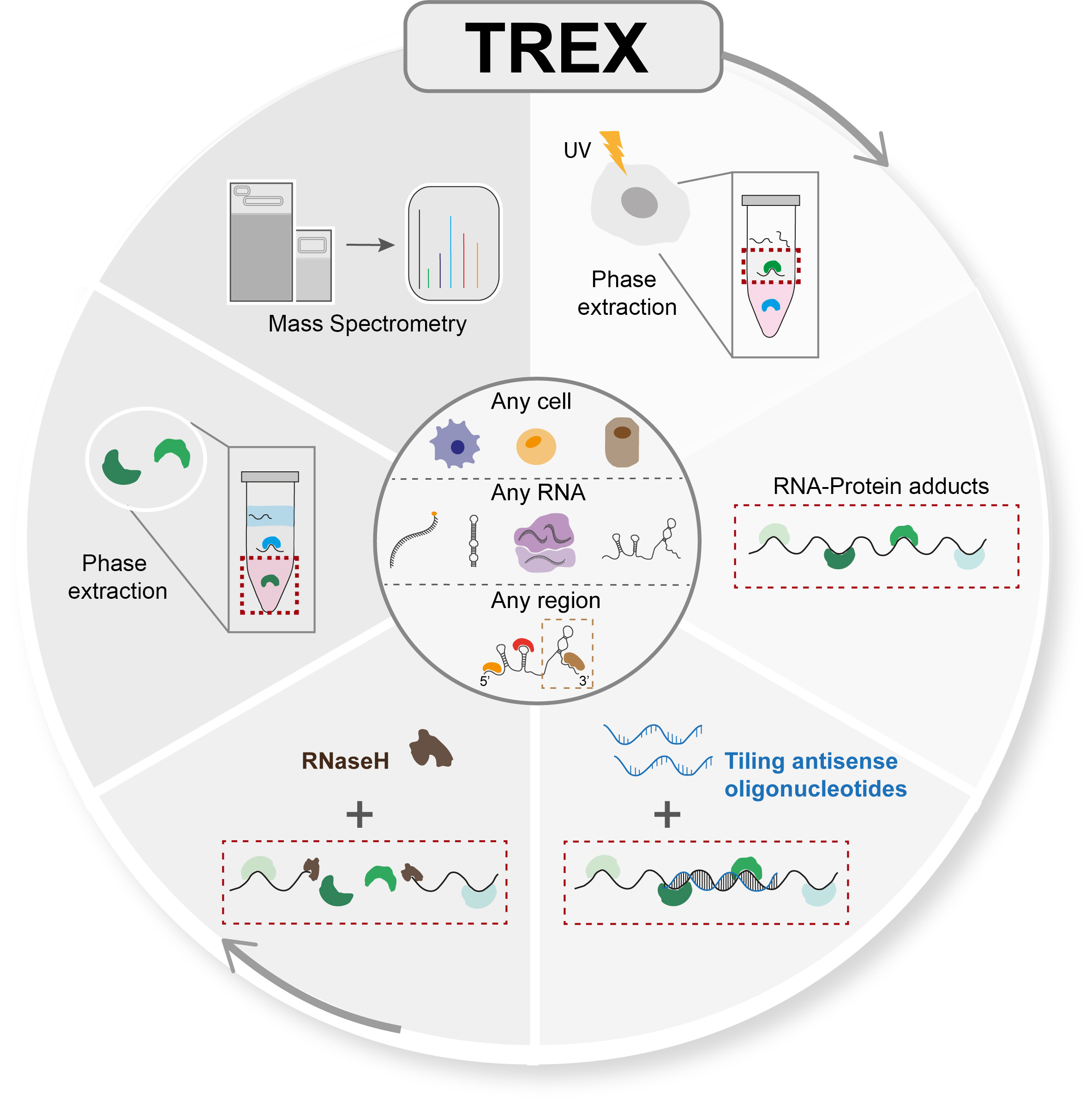
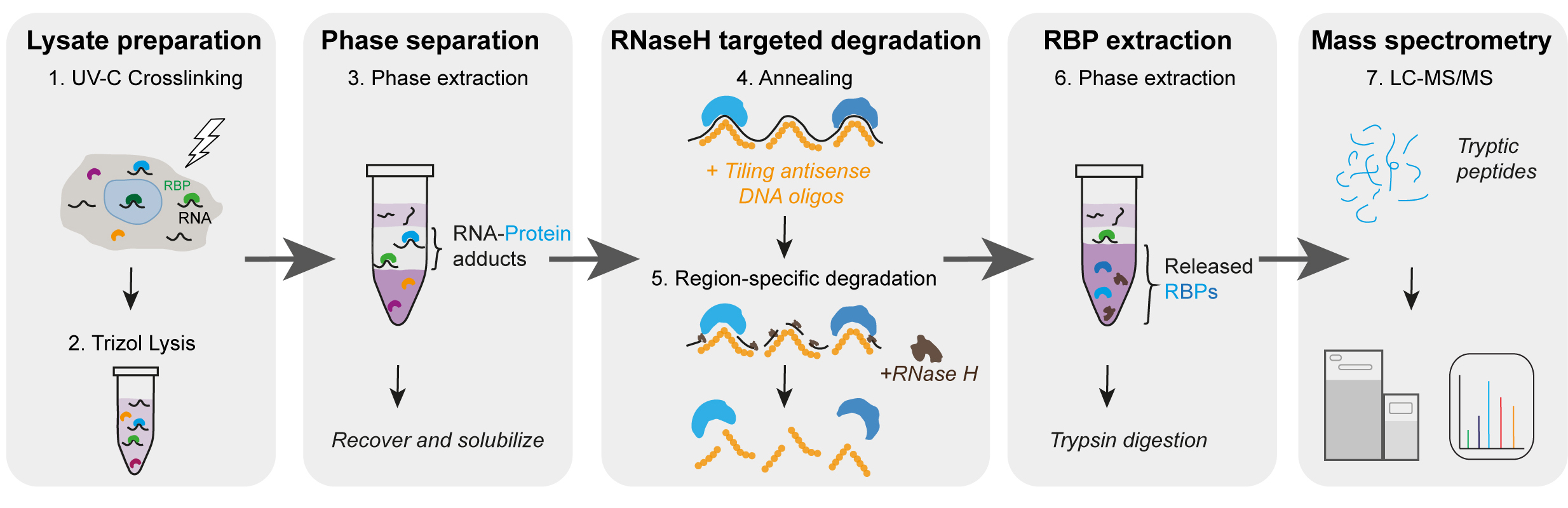
Brava Fede!(February 2024) Our TREX manuscript is now out in Nature Methods!
You can also read this short summary, describing the background to the work and what is special about TREX, now published as a Research Briefing:
https://rdcu.be/dy6ZI
If you are interesting in trying out TREX, Martin has also generated a detailed step-by-step protocol for it with lots of important advice and tips. You can check this out here on our website: https://www.mardakhehlab.info/resources/trex
(January 2024) Martin and Emilie talk at the RNA UK 2024 in Windermere!
Congratulations to Martin for delivering an excellent oral presentation at this year's RNA UK 2024 meeting in Windermere (25th to 28th of January). Martin talked about his recent work on TREX, a novel powerful method to reveal proteins that bind to specific RNA regions in living cells and under endogenous settings. Interested in knowing more? Check out his preprint on bioRxiv:
https://www.biorxiv.org/content/10.1101/2023.06.30.547259v1)
Also a big well done to Emilie for delivering an brilliant Flash talk on her project to reveal the causes and consequences of nucleolar changes in response to oncogenic RAS signalling! You both did the lab super proud!
(December 2023) Congratulations to Emilie for winning the Barts Cancer Institute 'PhD student Of The Year' Award!
Our lab concludes 2023 on a very high note, with our amazing PhD student, Emilie Alard, being selected as the BCI PhD student of the year!
Emilie has made very exciting discoveries during her project this year, as well as contributing to several other publications from our lab. But her hard work has not been restricted to our team! She has been generously supporting many other researchers across the different departments of the Institute, playing a crucial role in enabling them to design and carry out complex proteomics investigations. She has also played a pivotal role in the BCI PhD forum, organising numerous events for the PhD students, as well as being a key advocate for her peers.
Emilie received this prestigious award during our annual Institute's Christmas party, which was held at the historic Charterhouse square this year.
Well done Emilie! We are so proud for you!
Congratulations to Federica for delivering an excellent short talk at the Translation UK 2023 meeting, and winning the BEST TALK PRIZE! This was amongst some really excellent talks delivered this year, so a big well done to you Federica!
(July 2023) Check out the latest preprint from our lab entitled " TREX reveals proteins that bind to specific RNA regions in living cells" out now on bioRxiv!
This work was the result of a fun partnership and collaboration between our group and Lovorka Stojic's team, our neighbors here at Barts Cancer Institute! A big round of applause goes to our amazing Martin Dodel who did the bulk of TREX optimizations, and Giulia from Lo's lab who worked tirelessly beside Martin to do the key validations and analyses. Well done team!
You can read the work here on bioRxiv: https://doi.org/10.1101/2023.06.30.547259
Martin has also generated a detailed illustrated step-by-step protocol for TREX for anyone who may be interested in trying it out. You can find this on our website resource page here: https://www.mardakhehlab.info/resources/trex
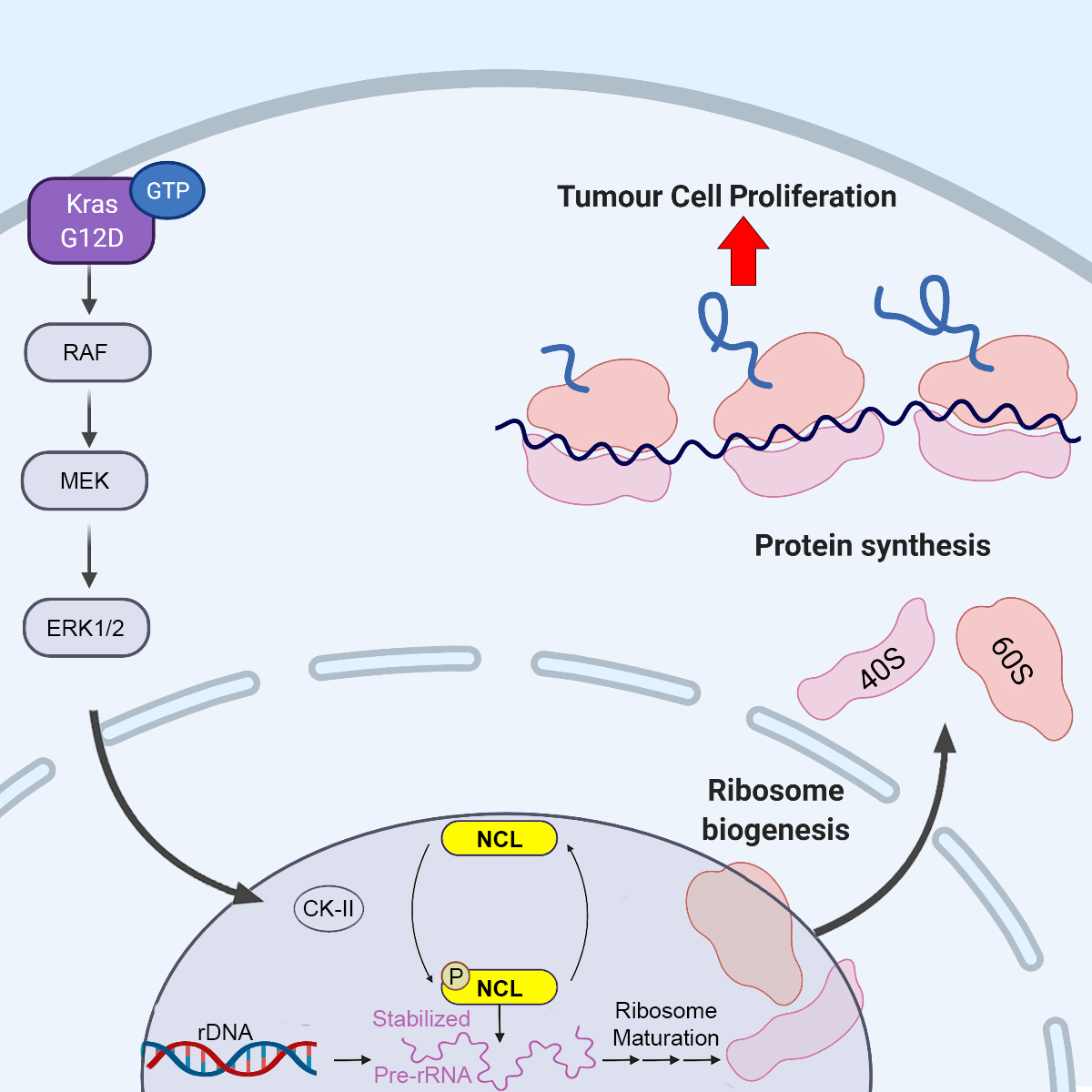
(April 2023) Our manuscript on oncogenic RAS regulation of ribosome biogenesis via nucelolin is finally out in EMBO Journal!
After nearly a year of hard work and addressing all the reviewers comments, our manuscript on RAS regulation of ribosome biogenesis through an ERK-CK2-nucleolin axis is finally out in EMBO Journal! Big congratulations to our recent PhD graduate Syahmi who initiated this project, Emilie who carried it over the finish line, and all other present and past Mardakheh team members (Martin, Fede, Maria, and Alina) who contributed to the work! Also, big thanks to Jernej Ule and Rupert Faraway, our amazing collaborators!
(March 2023) Congratulations to Elliott for successfully defending his PhD!
A big well done to Elliott for successfully defending his PhD on 24th of March! He becomes the second PhD student graduating from Mardakheh lab! Also many thanks to the fantastic viva examiners Prof. Marco Gerlinger (BCI) and Dr. Paul Huang (ICR) for conducting the viva! It was a pleasure to have you here and Elliott really enjoyed the experience!
Congratulations Dr Whittaker! I hope you enjoy all the special presents (and please don't finish them quickly :D)!
(March 2023) CRUK funded MBPhD project available in Mardakheh lab!
Are you a third year MBBS student at the Barts and the London School of Medicine and Dentistry? Are you interested in cancer research? Then look no more! As part of the CRUK City of London Centre (https://www.colcc.ac.uk/), we are offering an exciting MBPhD project (September 2023 start date) on understanding and exploiting the impact of Nucleophosmin1 (NPM1) mutations on modulation of ribosome synthesis in Acute Myeloid Leukaemia (AML).
NPM1 is an RNA-binding protein that plays a key role in organisation of the nucleoli, the subcellular sites of ribosome biogenesis. NPM1 mutations are found in >30% of all adult AML cases, and result in mislocalisation of the protein to the cytoplasm. However, the exact molecular impact of NPM1 mislocalisation on AML development is still unclear. This project aims to decipher the consequences of NPM1 mislocalisation on the nucleolus of AML cells, assessing how ribosome biogenesis is specifically modulated, and how such modulation may be therapeutically exploited in treatment of AML.
For more information on the project and how to apply, check out this page:
2023 MBPhD project Mardakheh – CRUK – City Of London Cancer Centre (colcc.ac.uk)
(January 2023) Congratulations to Martin for winning the Barts Cancer Institute 'Technician Of The Year' Award!
Massive congratulations to our technician Martin, who won the BCI technician of the year award. Martin not only has been a vital pillar of our lab, but also a tremendous help to others beyond our group. In the past year, Martin has enabled several proteomics collaborations with other BCI labs, trained many newcomers in our centre, and greatly helped with setting up our Institutes' brand new RNA suite. Martin has also been smashing it with his contributions to our labs papers and grants, as well as being an key contributor to an upcoming patent application from our team.
Thanks to all these amazing achievements, Martin received this award on the 27th of January from the BCI director, Professor Nick Lemoine (Pictured), during the BCI Burns night/New Year celebration Party, which was held at the iconic St Barts Great Hall.
Well done Martin! This one is so well deserved!
(December 2022) Federica wins the Best PhD Student Talk Prize at the South Coast RNA Network Meeting!
BIG Congratulations to Federica for delivering an excellent talk on her PhD work at the South Coast RNA Network Meeting, and winning the best PhD student talk prize! This years meeting was held at the University of Portsmouth, where Federica presented her new data on how LARP6 can recognise diverse functional groups RNAs inside the cell, as well as important insights on how these binding events may be regulating different pro-tumourigenic functions in mesenchymal cancers. Well done Fede!
(August 2022) Well done to Federica for presenting her work at the 6th biennial LARP society meeting in Toronto!
(March 2022) Congratulations to Dr. Syahmi for passing his PhD viva with flying colours!
(December 2021) Check out our latest preprint, out now on BioRxiv!
Our latest work describing how oncogenic RAS signaling modulates the RNA-bound proteome, is now out on bioRxiv: https://doi.org/10.1101/2021.12.16.472890
In this manuscript, we systematically profile changes in the RNA-bound proteome downstream of RAS oncogene, and discover a novel mechanism of ribosome biogenesis upregulation downstream of oncogenic RAS signalling, that is mediated via regulating the RNA-binding activity of Nucleolin (NCL).
A big well done to Syahmi who spearheaded this project, as well as Martin, Federica, Maria, and our MSc student Wanling, all of whom contributed to this study. Also big thanks goes to our fantastic collaborators, Rupert and Jernej for
their
critical support with regards to our NCL iCLIP analysis.
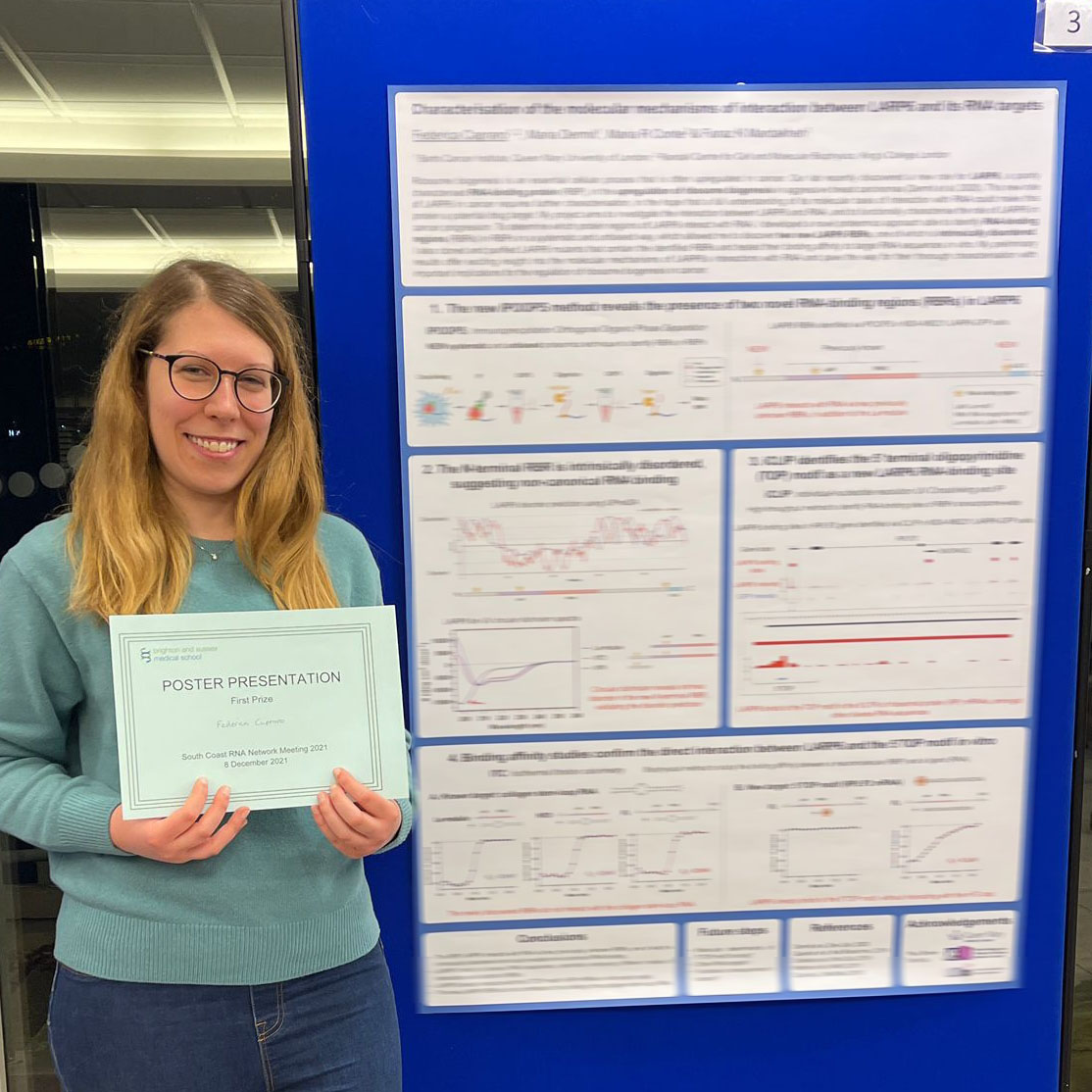
(December 2021) Best poster prize for Federica at the South Coast RNA Meeting!
(September 2021) We are hiring!
Are you interested in uncovering a novel mechanism of post-transcriptional dysregulation during cancer progression? We have a post-doctoral position available in our lab, so get in touch if you are curious!
https://webapps2.is.qmul.ac.uk/jobs/job.action?jobID=6220
If you would like to know more about this position, our lab, and working at our Institute before applying, just drop us an email (f.mardakheh[at]qmul.ac.uk).
(July 2021) Goodbye Maria!
Over the summer, we said goodbye to the first postdoc of Mardakheh's lab, the amazing Maria Dermit. Maria moved to the biotech sector to do some really cool science with stem cells at bit.bio. Even though we are very happy for you Maria, We will be really missing you (yes even your extremely messy bench!). We wish you all the best. Wherever you go to in your future endeavours, one thing for sure is that you will be achieving amazing things!
Adiós dama de T-Rex!
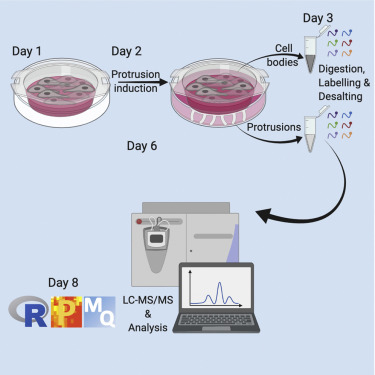
(April 2021) Check out our protocol for quantitative proteomics analysis of protrusions.
Interested in performing quantitative proteomics analysis of cell-protrusions like we did in our recent publication (Dermit et al., 2020)? If so, we have a detailed protocol for you, now published in STAR Protocols: https://doi.org/10.1016/j.xpro.2021.100462
Credit goes to Maria for preparing this extremely well-written and detailed protocol, which even has videos of the experiments shot on a body cam! Also check out her Github entry for all the data analysis pipelines you
need: https://demar01.github.io/protrusionproteome/index.html
(January 2021) We are hiring!
We have a fully funded CRUK PhD position available for the start of September 2021. Get in touch if you are interested in doing a PhD on RBPs and their role in pancreatic cancer. More details on the position, and how to apply can be found here:
https://www.bartscancer.london/about/vacancies/cancer-research-uk-phd-studentships/
(December 2020) BCI paper of the month award!
Our manuscript in Developmental Cell (Dermit et al., 2020) was chosen as the paper of the month by the Barts Cancer Institute's executive board. A very big well done to Maria and the team for their achievment!
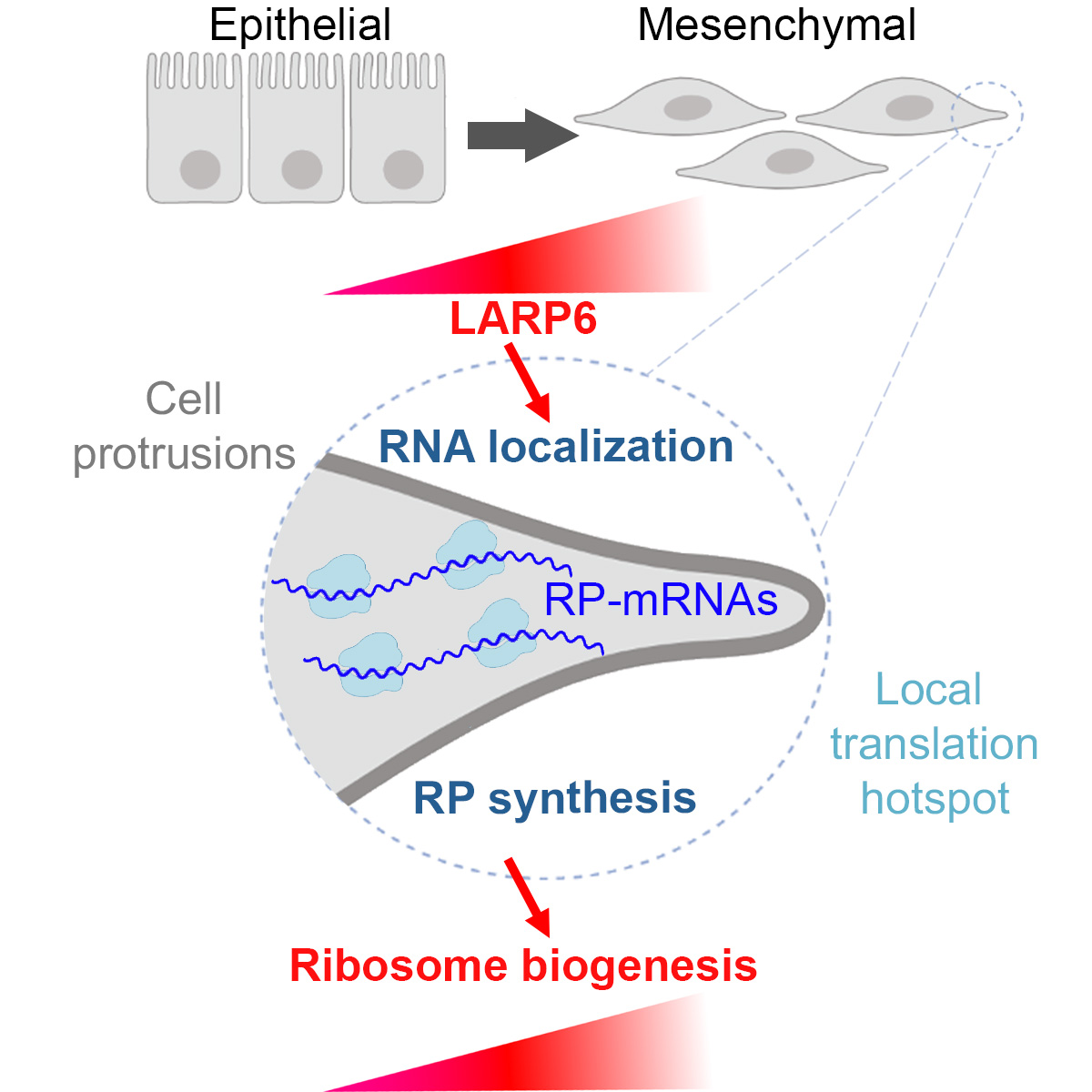
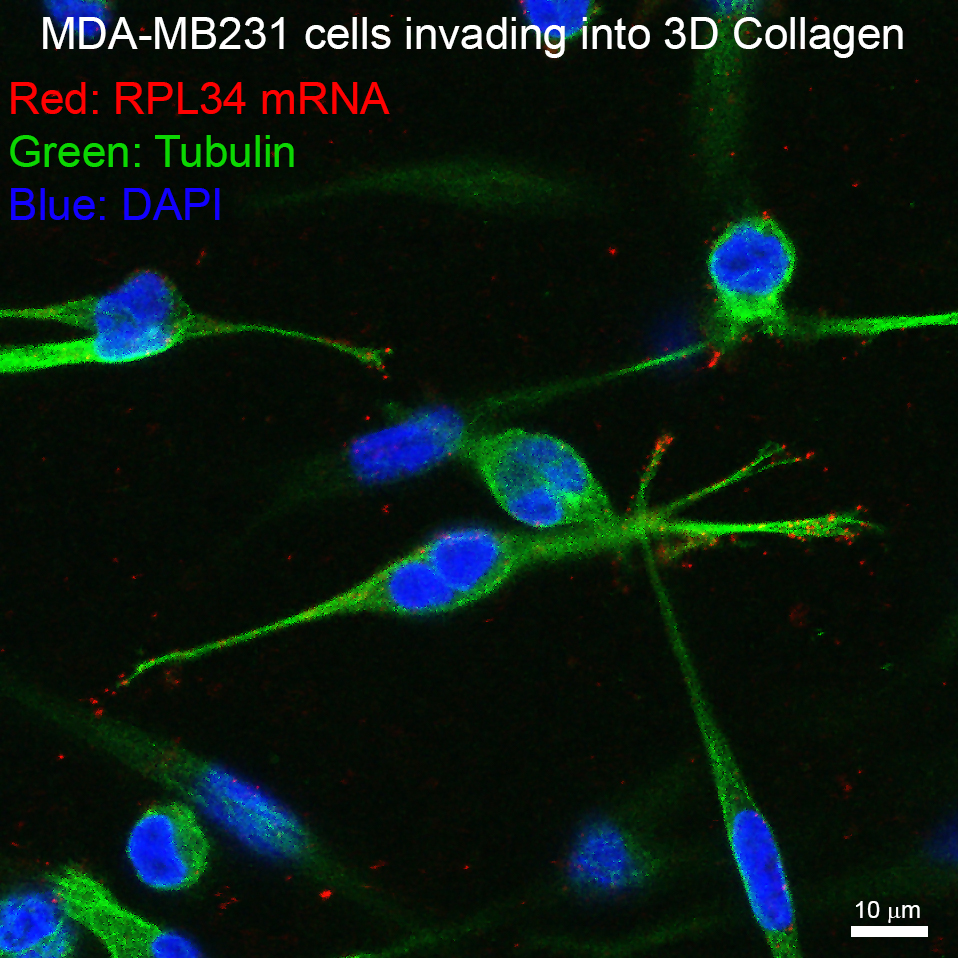
(November 2020) Our paper on LARP6 regulation of ribosome biogenesis by RNA localisation is out!
Almost 10 months after the initial submission, our paper on regulation of ribosome biogenesis by subcellular mRNA localisation is finally out in Developmental Cell! We are very proud of Maria for her great effort in completing the revisions despite huge challenges caused by the pandemic (e.g. 4 months of lockdown!). Many thanks to all the team members and collaborators who contributed to the work. We are also grateful to our funders MRC and Barts Charity for their generous support.
You can read the open-access paper here:
https://www.cell.com/developmental-cell/fulltext/S1534-5807(20)30796-6#%20
There is a wealth of multiomics data in the paper, including subcellular mRNA and protein distributions between protrusions and cell-bodies of 6 different human cell-lines. If you fancy exploring this data more, checking for instance how your mRNA or protein of interest is distributed between protrusions and cell-bodies in these cell-lines, you can also use this dashboard within our resource page:
(September 2020) Best poster prize for Syahmi!
Congratulations to Syahmi for winning the best poster prize at the Barts Cancer Institute's annual PhD day 2020. In his poster, Syahmi presented his findings on the importance of ribosome biogenesis for pancreatic Ductal Adenoarcinoma (PDAC) development, showing that addiction to mutant KRAS in PDAC depends on the KRAS ability to enhance ribosome biogenesis.
(July 2020) Back to the lab.
Roughly 3 ½ months passed since any of us lifted a pipette. But now the time has come to bring those old skills to the test. Even so only a few of us can be at the bench at once, we are doing our best to make up for the time lost. Continuing with where we left off, as well as working on new ideas which we have accumulated over the lock down.
(June 2020) Maria delivers a fantastic talk at BCI Thursday seminar series
Maria told us (remotely) about her recent work on LARP6 and regulation of ribosome biogenesis in migrating cells at BCI Thursday seminar series. Watch her full talk here (QMUL access only):
https://bcinet.qmcr.qmul.ac.uk/thursday-seminar-series-webinar-archive/
(December 2019) An update of our preprint is now available on bioRxiv
Check out the revised version of our preprint, now out on bioRxiv: https://www.biorxiv.org/content/10.1101/829739v2
(November 2019) Our lab's first ever preprint is now out on bioRxiv
Work spearheaded by our amazing postdoc, Maria Dermit, describes a novel mechanism by which mesenchymal-like migrating cells upregulate ribosomal protein synthesis and ribosome biogenesis, through RNA localization. Check out the manuscript here: https://www.biorxiv.org/content/10.1101/829739v1
(July 2019) Faraz presents at FASEB/EMBO RNA Localization and Local Translation Conference
Faraz and Maria attended this years' FASEB/EMBO RNA Localization and Local Translation conference in Snowmass, Colorado, where Faraz spoke about our soon to be published story on how spatial organisation of ribosomal protein mRNAs plays a crucial role in regulating ribosome biogenesis in migratory mesenchymal-like cells, and how this pathway is hijacked by highly aggressive cancer cells to enhance malignant growth and invasion.